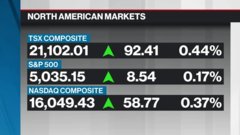
Nov 30, 2020
DeepMind's AlphaFold crosses threshold in solving protein riddle
, Bloomberg News

DeepMind Technologies Ltd.’s protein analysis project swept a major international competition, in a step toward making the tool more widely available to scientists studying new drugs and diseases.
AlphaFold, which uses artificial intelligence to predict the structure of proteins, beat other researchers in the latest Critical Assessment of Structure Prediction competition. The event started in 1994 and is held every two years to accelerate research on the topic.
DeepMind became a subsidiary of Google after a 2014 acquisition and is best known for its gamer AI, teaching itself to beat Atari video games and defeating world-renowned Go players like Lee Sedol. The company’s ambition has been to develop AI that can be applied to broader problems, and it’s so far created systems to make Google’s data centers more energy efficient, identify eye disease from scans and generate human-sounding speech.
The task of determining the structures of proteins has been described as a project akin to mapping the genome. Different folds in a protein determine how it will interact with other molecules, and understanding them has implications for discovering how new diseases like Covid-19 invade our cells, designing enzymes to break down pollutants and improving crop yields.
“These algorithms are now becoming strong enough and powerful enough to be applicable to scientific problems,” DeepMind Chief Executive Officer Demis Hassabis said in a call with reporters. After four years of development “we have a system that’s accurate enough to actually have biological significance and relevance for biological researchers.”
DeepMind is now looking into ways of offering scientists access to the AlphaFold system in a “scalable way,” Hassabis said.
“Citizen Science”
CASP scientists analyzed the shape of amino acid sequences for a set of about 100 proteins. Competitors were given the sequences, and charged with predicting their shape. AlphaFold’s assessment lined up almost perfectly with the CASP analysis for two-thirds of the proteins, compared to about 10% from the other teams, and better than what DeepMind’s tool achieved two years ago
Hassabis said his inspiration for AlphaFold came from “citizen science” attempts to find unknown protein structures, like Foldit, which presented amateur volunteers with the problem in the form of a puzzle. In its first two years, the human gamers proved to be surprisingly good at solving the riddles, and ended up discovering a structure that had baffled scientists and designing a new enzyme that was later confirmed in the lab.
“Determining a single protein structure often required years of experimental effort,” said Janet Thornton, director emeritus of the European Bioinformatics Institute and one of the pioneers of using computational approaches to understanding protein structure. “A better understanding of protein structures and the ability to predict them using a computer means a better understanding of life, evolution and, of course, human health and disease.”